JavaSwing实现的文本比较软件
2020-12-13 02:07
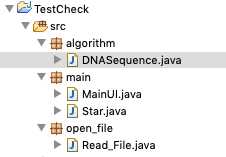
标签:ocs set 索引 因此 div NPU 参数 ade 右键 先看效果:截图1 截图2: 实现思路: 1、界面UI设计 2、功能点 : a 打开文件进行比较 b 粘贴内容进去比较 c 提示帮助 d 窗口可以任意拖动 3、文本比较算法 java类 : MainUI 类实现界面设计 Read_File 类实现文件读取 DNASequence 类文本比较算法 项目结构: 源代码: Star.java MainUI.java Read_File.java DNASequence.java 项目源代码已经上传到我的GitHub仓库 下载点击https://github.com/Wo-com/TextCheck 如果觉得不错的话 就在GitHub里面给我点亮 Star吧 JavaSwing实现的文本比较软件 标签:ocs set 索引 因此 div NPU 参数 ade 右键 原文地址:https://www.cnblogs.com/easyidea/p/11026060.html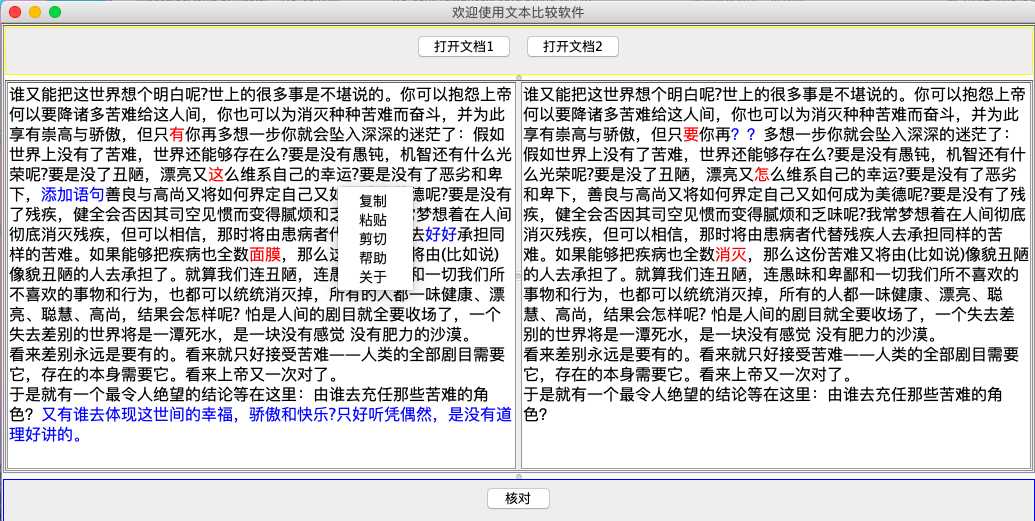


import javax.swing.text.BadLocationException;
import javax.swing.text.Document;
import javax.swing.text.SimpleAttributeSet;
import javax.swing.text.StyleConstants;
public class Star
{
public static void main(String args[]) throws BadLocationException
{
//初始化界面
Windowm windowm = new Windowm();
//属性设置
SimpleAttributeSet attrset = new SimpleAttributeSet();
//字体大小
StyleConstants.setFontSize(attrset,16);
//获取JTextPane对象
Document docs1=windowm.text1.getDocument();
//设置初次显示文本
docs1.insertString(docs1.getLength(), "手动输入或者选择文件打开", attrset);
Document docs2=windowm.text2.getDocument();
docs2.insertString(docs2.getLength(), "手动输入输入或者选择文间打开\n点击核对试试\n红色表示错误字符\n蓝色表示多余或缺失字符", attrset);
}
}


import algorithm.DNASequence;
import open_file.Read_File;
import java.awt.*;
import java.awt.event.ActionEvent;
import java.awt.event.ActionListener;
import java.awt.event.ComponentAdapter;
import java.awt.event.ComponentEvent;
import java.awt.event.MouseAdapter;
import java.awt.event.MouseEvent;
import java.io.File;
import javax.swing.*;
import javax.swing.text.Document;
import javax.swing.text.SimpleAttributeSet;
import javax.swing.text.StyleConstants;
class Windowm extends JFrame
{
String path1;//第一个文件目录
String path2;//第二个文件目录
String File1;//第一个文件
String File2;//第二个文件
int point;//保存当前活动窗口
private static final long serialVersionUID = 1L;
JPanel myPanel1 = new JPanel();//面板1.1
JPanel myPanel2 =new JPanel();//面板2.1
JPanel myPanel3 =new JPanel();//面板3
JPanel myPanel4 =new JPanel();//面板4
JTextPane text1=new JTextPane();
JTextPane text2=new JTextPane();
JButton bt1 = new JButton("打开文档1");
JButton bt2 = new JButton("打开文档2");
JButton bt3 = new JButton("核对");
JPopupMenu jm = new JPopupMenu();//右键菜单
JMenuItem copy = new JMenuItem("复制");//菜单项
JMenuItem path = new JMenuItem("粘贴");
JMenuItem cut = new JMenuItem("剪切");
JMenuItem help = new JMenuItem("帮助");
JMenuItem about = new JMenuItem("关于");
JScrollPane scro1=new JScrollPane(text1);//添加滚动条
JScrollPane scro2=new JScrollPane(text2);//添加滚动条
JSplitPane jSplitPane =new JSplitPane();//设定为拆分布局
JSplitPane jSplitPane2 =new JSplitPane();//设定为拆分布局
JSplitPane jSplitPane3 =new JSplitPane();//设定为拆分布局
public Windowm()
{
setVisible(true);
jm.add(copy);
jm.add(path);
jm.add(cut);
jm.add(help);
jm.add(about);
myPanel3.add(bt1);
myPanel3.add(bt2);
myPanel4.add(bt3);
this.setTitle("欢迎使用文本比较软件");
this.setBounds(100, 100, 600, 500);
jSplitPane.setContinuousLayout(true);//操作箭头,重绘图形
jSplitPane2.setContinuousLayout(true);//操作箭头,重绘图形
jSplitPane3.setContinuousLayout(true);//操作箭头,重绘图形
jSplitPane2.setOrientation(JSplitPane.VERTICAL_SPLIT);//垂直方向
jSplitPane.setOrientation(JSplitPane.HORIZONTAL_SPLIT);//水平方向
jSplitPane3.setOrientation(JSplitPane.VERTICAL_SPLIT);//垂直方向
myPanel1.setBorder(BorderFactory.createLineBorder(Color.green));
myPanel2.setBorder(BorderFactory.createLineBorder(Color.red));
myPanel3.setBorder(BorderFactory.createLineBorder(Color.yellow));
myPanel4.setBorder(BorderFactory.createLineBorder(Color.blue));
jSplitPane.setLeftComponent(scro1);//左右布局中添加组件 ,面板1
jSplitPane.setRightComponent(scro2);//左右布局中添加组件 ,面板2
jSplitPane2.setTopComponent(myPanel3);//上下布局中添加组件 ,面板1
jSplitPane2.setBottomComponent(jSplitPane);//上下布局中添加组件 ,面板1
jSplitPane3.setTopComponent(jSplitPane2);
jSplitPane3.setBottomComponent(myPanel4);
jSplitPane.setDividerSize(5);//设置分割线的宽度
jSplitPane2.setDividerSize(5);//设置分割线的宽度
jSplitPane3.setDividerSize(5);//设置分割线的宽度
setContentPane(jSplitPane3);//设置为父模块
setDefaultCloseOperation(JFrame.EXIT_ON_CLOSE);
copy.addActionListener(new ActionListener()//窗口监听
{
public void actionPerformed(ActionEvent e4)//菜单项
{
try{
text1.copy();
text2.copy();
}catch(Exception e1){
}
}
}
);
path.addActionListener(new ActionListener()//窗口监听
{
public void actionPerformed(ActionEvent e4)//菜单项
{
try{
if(point==1)//由于有两个窗口,因此设计point来确定粘贴在某个窗口
text1.paste();
else
text2.paste();
}catch(Exception e1){
}
}
}
);
cut.addActionListener(new ActionListener()//窗口监听
{
public void actionPerformed(ActionEvent e4)//菜单项
{
try{
text1.cut();
text2.cut();
}catch(Exception e1){
}
}
}
);
help.addActionListener(new ActionListener()//窗口监听
{
public void actionPerformed(ActionEvent e4)//菜单项
{
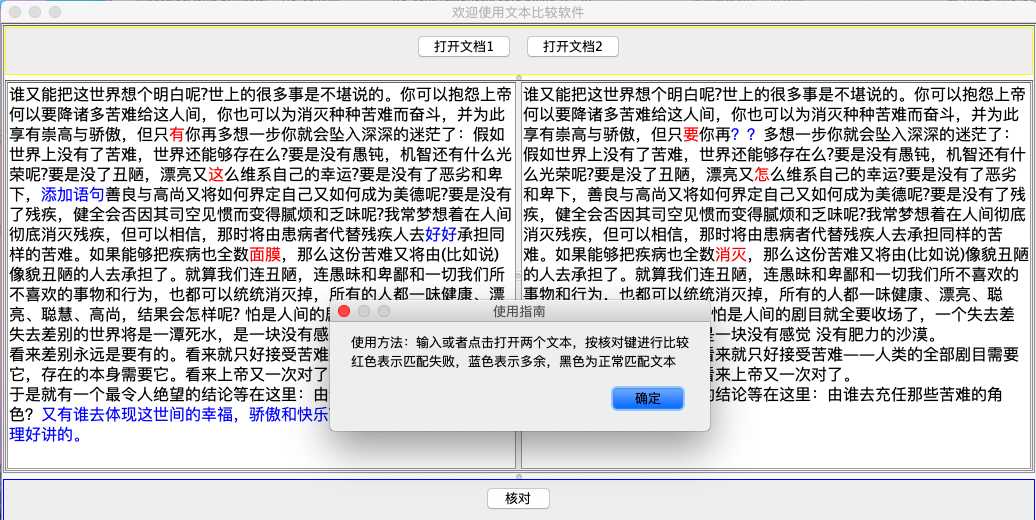
JOptionPane.showMessageDialog(null,"使用方法:输入或者点击打开两个文本,按核对键进行比较\n红色表示匹配失败,蓝色表示多余,黑色为正常匹配文本","使用指南",JOptionPane.PLAIN_MESSAGE);
}
}
);
about.addActionListener(new ActionListener()//窗口监听
{
public void actionPerformed(ActionEvent e4)//菜单项
{
JOptionPane.showMessageDialog(null,"原创:将军\nQQ 2910001378@qq.com \n人生苦短,欢迎转载","将军原创",JOptionPane.PLAIN_MESSAGE);
}
}
);
text1.addMouseListener(new MouseAdapter() {
public void mouseClicked(MouseEvent e) {
if (e.getButton() == MouseEvent.BUTTON3) {
jm.show(text1, e.getX(), e.getY()); // 弹出菜单
point=1;
}
}
});
text2.addMouseListener(new MouseAdapter() {
public void mouseClicked(MouseEvent e) {
if (e.getButton() == MouseEvent.BUTTON3) {
jm.show(text2, e.getX(), e.getY()); // 弹出菜单
point=2;
}
}
});
jSplitPane.addComponentListener(new ComponentAdapter() {//拖动窗口监听
public void componentResized(ComponentEvent e) {
jSplitPane.setDividerLocation(jSplitPane3.getWidth()/2-7);//设置第一条宽度
}
});
jSplitPane2.setDividerLocation(50);//设定分割线的距离左边的位置
jSplitPane3.addComponentListener(new ComponentAdapter() {//拖动窗口监听
public void componentResized(ComponentEvent e) {
jSplitPane3.setDividerLocation(jSplitPane3.getHeight()-50);//设置第三条高度
}
});
bt1.addActionListener(new ActionListener()//窗口监听
{
public void actionPerformed(ActionEvent e4)//菜单项
{
try{
text1.setText("");
JFileChooser jfc=new JFileChooser();
jfc.setFileSelectionMode(JFileChooser.FILES_AND_DIRECTORIES );
jfc.showDialog(new JLabel(), "选择");
File file=jfc.getSelectedFile();
path1=file.getAbsolutePath();//获取文件绝对地址
new Read_File(path1);
File1= Read_File.getFile();
SimpleAttributeSet attrset = new SimpleAttributeSet();
StyleConstants.setFontSize(attrset,16);//设置字号
Document docs=text1.getDocument();
docs.insertString(docs.getLength(), File1, attrset);
}catch(Exception e1){
}
}
}
);
bt2.addActionListener(new ActionListener()//窗口监听
{
public void actionPerformed(ActionEvent e4)//菜单项
{
try{
text2.setText("");
JFileChooser jfc=new JFileChooser();
jfc.setFileSelectionMode(JFileChooser.FILES_AND_DIRECTORIES );
jfc.showDialog(new JLabel(), "选择");
File file=jfc.getSelectedFile();
path2=file.getAbsolutePath();//获取文件绝对地址
new Read_File(path2);
File2= Read_File.getFile();
SimpleAttributeSet attrset = new SimpleAttributeSet();
StyleConstants.setFontSize(attrset,16);//设置字号
Document docs=text2.getDocument();
docs.insertString(docs.getLength(), File2, attrset);
}catch(Exception e1){
System.out.println("选择文件出错");
}
}
}
);
bt3.addActionListener(new ActionListener()//窗口监听
{
public void actionPerformed(ActionEvent e4)//菜单项
{
try{
String dnas1;//算法处理之后的字符串1
String dnas2;//算法处理之后的字符串2
String jtp1;//JTextpane的内容1
String jtp2;//JTextpane的内容2
int len=0; //处理后的字符串长度
jtp1=text1.getText();//获取窗口文本
jtp2=text2.getText();
text1.setText("");//清空之前内容
text2.setText("");
Document docs1=text1.getDocument();
Document docs2=text2.getDocument();
DNASequence dna=new DNASequence(jtp1,jtp2);//通过构造方法传递参数
dna.runAnalysis();
dna.traceback();
dnas1=dna.getString1();//获取处理后的字符串
dnas2=dna.getString2();
char[] s = dnas1.toCharArray();//字符串转Char数组
char[] p = dnas2.toCharArray();
len=dnas1.length();
SimpleAttributeSet set2 = new SimpleAttributeSet();//设置一个属性
StyleConstants.setFontSize(set2,16);//设置字号
for(int i=0;i


import java.io.BufferedReader;
import java.io.FileInputStream;
import java.io.InputStreamReader;
public class Read_File {
static String fileName;
public Read_File(String str){
fileName=str;
}
public static String getFile(){
BufferedReader br = null;
StringBuffer sb = null;
try {
br = new BufferedReader(new InputStreamReader(new FileInputStream(fileName),"utf-8")); //这里可以控制编码
sb = new StringBuffer();
String line = null;
while((line = br.readLine()) != null) {
sb.append(line);
}
} catch (Exception e) {
e.printStackTrace();
} finally {
try {
br.close();
} catch (Exception e) {
e.printStackTrace();
}
}
String data = new String(sb); //StringBuffer ==> String
System.out.println("数据为==> " + data);
return data;
}
// public static void main(String[] args) {
// new Read_File("1");
// }
}


//https://codereview.stackexchange.com/questions/182601/optimizing-needleman-wunsch-algorithm-in-java
public class DNASequence {
protected final String seq_1, seq_2; //要分析的两个序列
public int alignmentScore; //使用Needleman-Wunsch算法
protected Node[][] matrix; //存储分数和缩进
protected final int matchScore, mismatchScore, indel;
//用于打印DNA序列分析的字符串
String top = ""; // Sequence 1
String bottom = ""; // Sequence 2
public DNASequence(String s1, String s2) {
//我使用一个■作为缓冲,这样序列字符串正确对齐
// 在矩阵中使用缩进和分数。
ScoreScheme s = new ScoreScheme(2, -1, -2);
seq_1 = "\u25A0" + s1;
seq_2 = "\u25A0" + s2;
matchScore = s.matchScore;
mismatchScore = s.mismatchScore;
indel = s.indel;
matrix = new Node[seq_1.length()][seq_2.length()];
for (int i = 0; i )
matrix[i][0] = new Node(i * indel, i, 0);
for (int i = 0; i )
matrix[0][i] = new Node(i * indel, 0, i);
}
//辅助方法,帮助决定使用哪种匹配/不匹配得分。
protected int score(int i, int j) {
if (seq_1.charAt(i) == seq_2.charAt(j))
return matchScore;
else
return mismatchScore;
}
//在本地级别上实现Needleman-Wunsch algo的Helper方法。
protected Node match(int i, int j) {
Node s1,s2,s3;
s1 = new Node(matrix[i-1][j-1].score + score(i, j), i, j);
s2 = new Node(matrix[i-1][j].score + indel, i, j);
s3 = new Node(matrix[i][j-1].score + indel, i, j);
Node largest = new Node(Math.max(s1.score, Math.max(s2.score, s3.score)), i, j);
if (s1.compareTo(largest) == 0)
largest.prev = matrix[i-1][j-1];
else if(s2.compareTo(largest) == 0)
largest.prev = matrix[i-1][j];
else
largest.prev = matrix[i][j-1];
return largest;
}
public Node runAnalysis() {
for (int i = 1; i ) {
for (int j = 1; j ){
matrix[i][j] = match(i, j);
}
}
alignmentScore = matrix[seq_1.length()-1][seq_2.length()-1].score;
return matrix[seq_1.length()-1][seq_2.length()-1];
}
//辅助方法,逐步构建分析结果。它将返回
//“尾巴”,因为我们可能还需要做一些工作。
protected Node traceHelper(Node curr) {
while (curr.prev != null) {
if (curr.i - curr.prev.i == 1 && curr.j - curr.prev.j == 1){ // If the path leads diagonal
boolean x = seq_1.charAt(curr.i) == seq_2.charAt(curr.j) ? true : false;
if(x){
top = seq_1.charAt(curr.i) +top;
bottom = seq_2.charAt(curr.j) +bottom;
}else{
top = seq_1.charAt(curr.i) + top;
bottom = seq_2.charAt(curr.j) + bottom;
}
}else if (curr.i - curr.prev.i == 1){ //如果这条路通向山顶
top = seq_1.charAt(curr.i) + top;
bottom = "-" + bottom; //如果这条路通向左边
}else if (curr.j - curr.prev.j == 1){
top = "-" + top;
bottom = seq_2.charAt(curr.j) + bottom;
}
curr = curr.prev;
}
return curr;
}
//从矩阵的最后一个节点回溯到第一个索引节点。
public void traceback() {
Node curr = matrix[seq_1.length()-1][seq_2.length()-1];
curr = traceHelper(curr);
while (curr.i != 0 || curr.j != 0) {
if (curr.i != 0 && curr.j == 0){
curr.prev = matrix[curr.i-1][curr.j];
curr = traceHelper(curr);
}else if (curr.i == 0 && curr.j != 0) {
curr.prev = matrix[curr.i][curr.j-1];
curr = traceHelper(curr);
}
}
//打印DNA序列分析
// System.out.println(top);
// System.out.println(bottom);
}
public String getString1(){
return top;
}
public String getString2(){
return bottom;
}
}
class Node implements Comparable